Phenoarch platform - Cleaning procedure - Curve level - CARBayesST library
I.Sanchez
septembre 04, 2020
CARBayesSTReport.RmdObjective
Detection of outliers in time courses of an experiment in PhenoArch greenhouse. In this vignette, we use a toy data set of the openSilexStatR library (anonymized real data set).
Please, have a look on the names of the columns of the input data set. The function is not generic and needs specific columns names in the input data set:
- Line: the x-coordinate in the lattice (numeric)
- Position: the y-coordinate in the lattice (numeric)
- Ref: a unique identifiant by pot, repetition and alias
- scenario: the scenario applied to the experiment
- genotypeAlias: the used genotypes in the experiment
The example is conducted on plantHeight parameter, we modelise the plantHeight taking into account the temporality (thermal time) and the spatiality (lattice structure) with a bayesian spatio-temporal ANOVA model [3].
To detect outlier points, we retrieve the standardised residuals computed by the model and a point is an outlier if abs(res) > threshold (here threshold==4). the user will be able to change this threshold.
We produced the output of the model as well as some graphics.
Import of data
The input dataset must contain some predefined columns, have a look to the structure of mydata:
library(lubridate) library(dplyr) library(openSilexStatR) myReport<-substr(now(),1,10) mydata<-plant1 str(mydata)
## 'data.frame': 47022 obs. of 14 variables:
## $ Ref : Factor w/ 1680 levels "manip1_10_10_WW",..: 131 131 131 131 131 131 131 131 131 131 ...
## $ experimentAlias: Factor w/ 1 level "manip1": 1 1 1 1 1 1 1 1 1 1 ...
## $ Day : Factor w/ 42 levels "2013-02-01","2013-02-02",..: 3 4 5 6 7 9 9 10 11 12 ...
## $ potAlias : int 1 1 1 1 1 1 1 1 1 1 ...
## $ scenario : Factor w/ 2 levels "WD","WW": 2 2 2 2 2 2 2 2 2 2 ...
## $ genotypeAlias : Factor w/ 274 levels "11430_H","A310_H",..: 165 165 165 165 165 165 165 165 165 165 ...
## $ repetition : int 1 1 1 1 1 1 1 1 1 1 ...
## $ Line : int 1 1 1 1 1 1 1 1 1 1 ...
## $ Position : int 1 1 1 1 1 1 1 1 1 1 ...
## $ thermalTime : num 1.29 2.65 3.98 5.32 6.66 ...
## $ plantHeight : num 140 151 213 239 271 ...
## $ leafArea : num 0.018 0.019 0.0208 0.0222 0.0235 ...
## $ biovolume : num 0.253 0.62 1.201 1.68 3.396 ...
## $ Repsce : Factor w/ 15 levels "1_WD","1_WW",..: 2 2 2 2 2 2 2 2 2 2 ...CARBayesST library
A bayesian approach to model these data where the spatio-temporal structure is modelled via sets of autocorrelated random effets. Conditional autoregressive (CAR) priors and spatio-temporal extensions thereof are typically assigned to these random effects to capture the autocorrelation, which are special cases of a Gaussian Markov random Filed (GMRF) [3].
ST.CARanova() decomposes the spatio-temporal variation into 3 components:
- an overall spatial effect common to all time periods
- an overall temporal trend common to all spatial units
- a set of independent space-time interactions
we can add others factors (here the scenario (quali) …)
The spatio-temporal auto-correlation is modelled by a common set of spatial random effect and a common set of temporal random effects and both are modelled by the CAR prior (Conditional AutoRegressive).
Description of the results’s table:
- Median: posterior median estimation
- 2.5 and 97.5%: 95% credible interval
- Geweke.diag: the convergence diagnostic implemented in the coda package. takes form of a Z-score, so the convergence is suggested by the statistic being within the range (-1.96; 1.96)
Model fit criteria
- DIC: deviance information criterion
- p.d: its corresponding estimated effective number of parameters
- WAIC: the Watanabe-Akaike information criterion
- p.w: its corresponding estimated number of effective parameters
- LMPL: the log marginal predictive likelihood
In model’s comparison, the best fitting model is the one that minimises the DIC and WAIC but maximises the LMPL.
As the bayesian model can take a while, in this example, we initialise burnin=10 and n.sample=110 - not enough in real analysis!
# We suppress observations with missing data in time variable (here thermalTime) mydata<-filter(mydata,!is.na(mydata$thermalTime)) model<-fitCARBayesST(datain=mydata,xvar="thermalTime",trait="plantHeight",k=2, graphDist=TRUE,burnin=10,n.sample=110, formulaModel=as.formula(plantHeight~scenario+genotypeAlias),typeModel="anova",verbose=TRUE)
## [1] 22.35558
## Setting up the model.
## Generating 100 post burnin and thinned (if requested) samples.
##
|
| | 0%
|
|= | 1%
|
|= | 2%
|
|== | 3%
|
|=== | 4%
|
|==== | 5%
|
|==== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 20%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 23%
|
|================= | 24%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 30%
|
|====================== | 31%
|
|====================== | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 40%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 47%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|======================================= | 55%
|
|======================================= | 56%
|
|======================================== | 57%
|
|========================================= | 58%
|
|========================================= | 59%
|
|========================================== | 60%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 63%
|
|============================================= | 64%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 67%
|
|================================================ | 68%
|
|================================================ | 69%
|
|================================================= | 70%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 100%
## Summarising results.
## Finished in 138.8 seconds.
## used (Mb) gc trigger (Mb) max used (Mb)
## Ncells 3642614 194.6 5520292 294.9 3642614 194.6
## Vcells 67027796 511.4 212701377 1622.8 67027796 511.4# print the result of the bayesian modelling printCARBayesST(modelin=model[[1]])
## Median 2.5% 97.5% Geweke.diag
## (Intercept) 789.1258 769.8793 809.8913 -2.3
## scenarioWW 5.0597 0.8237 8.4879 -1.8
## genotypeAliasA310_H -31.6065 -53.5133 -10.8240 -0.7
## genotypeAliasA347 -291.4734 -346.1374 -259.5544 2.0
## genotypeAliasA347_H -9.4935 -35.1732 32.5818 3.7
## genotypeAliasA374_H 62.2399 24.6144 92.4539 0.6
## genotypeAliasA375_H 94.3326 76.7937 117.1912 -3.6
## genotypeAliasA3_H 84.8671 53.7670 100.8658 1.4
## genotypeAliasA554_H -71.3099 -101.7732 -38.5134 1.8
## genotypeAliasAS5707_H 38.1778 3.7827 67.4886 0.6
## genotypeAliasB100_H 23.8927 -10.7963 58.7000 3.3
## genotypeAliasB104_H -36.1082 -76.1128 2.5110 0.0
## genotypeAliasB105_H -45.0814 -75.0145 -18.0219 0.0
## genotypeAliasB106_H 40.6225 5.8670 82.8446 -0.2
## genotypeAliasB107_H -65.6608 -100.2115 -40.4276 -0.3
## genotypeAliasB108_H 37.3071 -26.5343 69.7652 -12.3
## genotypeAliasB109_H 27.1687 4.9619 59.8399 -2.3
## genotypeAliasB110_H -34.1206 -71.1262 5.3274 0.4
## genotypeAliasB113_H 25.3901 -28.1445 64.6275 1.4
## genotypeAliasB14a_H -87.2072 -111.0793 -49.4009 5.6
## genotypeAliasB37_H -44.5693 -86.6378 -15.8982 0.5
## genotypeAliasB73 -156.9179 -196.9022 -110.0927 1.6
## genotypeAliasB73_H 16.5924 -18.5922 50.5890 2.3
## genotypeAliasB73_SILK -13.6384 -39.6926 23.5033 -1.3
## genotypeAliasB84_H -0.5810 -49.5477 38.6836 -1.3
## genotypeAliasB89_H -31.2775 -80.3974 5.0282 -3.8
## genotypeAliasB97_H 90.6655 40.2888 123.8072 0.3
## genotypeAliasB98_H -85.2713 -106.2585 -53.9576 0.2
## genotypeAliasC103_H 5.0336 -37.4723 27.2470 1.1
## genotypeAliasCO109_H -1.6207 -29.9578 27.8305 0.1
## genotypeAliasCR1Ht_H 10.9217 -16.5854 53.2746 0.5
## genotypeAliasD09_H 29.3918 -15.9286 49.9769 -7.5
## genotypeAliasDE811_H -90.6326 -108.4585 -59.8798 -0.1
## genotypeAliasDK2MA22_H 81.5465 58.6133 106.6624 -0.1
## genotypeAliasDK4676A_H -36.0684 -66.4866 13.7882 -0.8
## genotypeAliasDK78010_H -61.9133 -86.8355 -19.3594 -0.2
## genotypeAliasDK78371A_H 1.7957 -21.5624 15.6482 1.7
## genotypeAliasDKFAPW_H -25.0512 -45.8046 11.8085 -1.7
## genotypeAliasDKFBHJ_H 47.5936 16.6545 84.8306 0.3
## genotypeAliasDKIBO2_H -7.8186 -29.5641 16.4004 3.4
## genotypeAliasDKMBST_H 102.3405 73.5257 129.6724 -2.5
## genotypeAliasEA1027_H 52.0809 5.1974 83.5798 -2.1
## genotypeAliasEA1163_H -5.2095 -42.5073 32.8080 -3.0
## genotypeAliasEA3076_H -18.9045 -53.2195 11.2809 -1.7
## genotypeAliasEC136_H -18.8040 -41.5599 -1.4261 -0.8
## genotypeAliasEC140_H -73.8020 -98.5111 -53.7462 -3.8
## genotypeAliasEC151_H -68.5053 -125.2913 -43.3861 -3.5
## genotypeAliasEC169_H -16.3148 -45.7585 4.7795 0.9
## genotypeAliasEC175_H -53.2710 -83.6437 -36.5791 -1.8
## genotypeAliasEC232_H -20.0695 -70.0898 -2.3298 -1.3
## genotypeAliasEC242C_H 26.9978 -32.6042 64.7961 2.7
## genotypeAliasEC334_H -76.6495 -102.1347 -52.0159 0.4
## genotypeAliasEP10_H 46.6883 19.3753 76.0120 0.4
## genotypeAliasEP2008-18_H 33.9042 -6.1957 53.0412 1.2
## genotypeAliasEP2008-22_H 11.4261 -24.6468 35.9063 2.4
## genotypeAliasEP29_H -59.8603 -81.1789 -32.7272 -1.1
## genotypeAliasEP51_H -33.1826 -90.4161 9.5249 -2.8
## genotypeAliasEP52_H 7.5433 -7.7806 34.5417 -1.4
## genotypeAliasEP55_H -132.6336 -158.0428 -108.6033 4.1
## genotypeAliasEP67_H 0.6530 -25.7210 51.2536 -0.7
## genotypeAliasEP72_H 27.4721 -14.9591 49.0887 0.7
## genotypeAliasEP77_H 99.5765 64.2801 136.7359 0.3
## genotypeAliasEZ11A_H -39.2953 -86.3965 -13.1856 1.5
## genotypeAliasEZ18_H 26.0612 1.0650 57.3930 -0.6
## genotypeAliasEZ31_H 60.4100 21.1816 96.4638 -1.5
## genotypeAliasEZ34_H -48.1943 -79.7719 -1.5748 -1.6
## genotypeAliasEZ35_H -14.6472 -45.5773 22.2358 -4.1
## genotypeAliasEZ36_H -45.0150 -83.9646 1.2946 5.1
## genotypeAliasEZ37_H -32.2826 -55.0748 -4.7252 0.0
## genotypeAliasEZ38_H -22.0290 -53.7416 7.4692 -0.2
## genotypeAliasEZ40_H -70.9698 -112.1738 -43.9648 -1.9
## genotypeAliasEZ42_H -52.2353 -79.6463 -23.0109 -0.7
## genotypeAliasEZ47_H 62.6740 24.0514 87.8071 -0.1
## genotypeAliasEZ48_H 14.0183 -22.0985 48.2011 -0.7
## genotypeAliasEZ53_H -74.1028 -102.6507 -30.1623 -3.9
## genotypeAliasEZ5_H 26.7060 6.9257 61.0339 0.7
## genotypeAliasF04401_H -31.4035 -53.2366 -4.4456 0.4
## genotypeAliasF04402_H -77.5216 -112.2109 -54.3325 -0.4
## genotypeAliasF04701_H -28.7023 -58.0293 -0.0631 -1.5
## genotypeAliasF04702_H 27.6270 0.7867 51.2064 -2.5
## genotypeAliasF05101_H 80.4315 42.9681 111.0378 0.5
## genotypeAliasF05404_H 46.6131 17.8816 70.9081 -1.0
## genotypeAliasF1808_H -9.8075 -43.2317 9.7917 4.6
## genotypeAliasF1890_H 41.2684 -0.7654 79.9087 -2.5
## genotypeAliasF218_H -33.0417 -69.6055 -6.3558 -4.7
## genotypeAliasF252_H 70.4609 22.6894 93.5266 -5.4
## genotypeAliasF353 -124.3517 -198.2351 -86.9601 1.8
## genotypeAliasF353_H 99.5496 68.3792 140.9172 3.0
## genotypeAliasF354_H 17.5394 -30.6944 41.7917 1.9
## genotypeAliasF584_H -39.1503 -80.8775 -11.1845 -1.7
## genotypeAliasF608_H 34.5193 -0.7889 78.4127 -2.9
## genotypeAliasF618_H -10.0301 -40.8263 28.4285 0.4
## genotypeAliasF7001_H -20.3955 -56.5213 12.8960 -0.3
## genotypeAliasF7019_H 34.3594 -1.1819 59.3596 -0.9
## genotypeAliasF7025_H -48.6501 -78.0695 -11.1032 0.4
## genotypeAliasF7028_H 38.9339 17.0080 65.9701 0.9
## genotypeAliasF7057_H 5.2002 -20.8543 32.6682 1.9
## genotypeAliasF7058_H 21.3338 -36.0228 53.9361 -2.0
## genotypeAliasF7081_H 53.5041 17.0029 86.4948 0.9
## genotypeAliasF7082_H 45.9980 9.0246 72.3844 1.0
## genotypeAliasF712_H 75.2930 38.2709 103.3222 -1.6
## genotypeAliasF748_H -56.4360 -108.0607 -31.7268 -3.7
## genotypeAliasF752_H -22.8158 -56.9289 28.5490 -2.5
## genotypeAliasF816_H 27.3965 0.4608 52.8988 -2.4
## genotypeAliasF838_H -22.8282 -62.5700 14.6101 0.3
## genotypeAliasF874_H 29.7280 -36.0930 65.5741 -5.0
## genotypeAliasF888_H 50.5120 6.4823 92.5947 0.1
## genotypeAliasF894_H -85.6167 -117.2810 -39.4607 -0.5
## genotypeAliasF908_H -37.8413 -66.3360 -15.1863 1.1
## genotypeAliasF912_H 49.6091 21.6496 69.8835 0.3
## genotypeAliasF918_H 11.9352 -17.3708 41.2382 -1.1
## genotypeAliasF922_H 152.2907 111.4490 183.0062 -4.6
## genotypeAliasF924 -213.2465 -269.3964 -182.3181 2.5
## genotypeAliasF924_H 40.2170 -0.5559 80.3479 5.7
## genotypeAliasF924_SILK -27.5185 -60.0102 -0.7748 -1.3
## genotypeAliasF98902_H 96.6630 42.7745 152.4120 -0.7
## genotypeAliasFP1_H 86.6334 36.6928 143.2701 -2.2
## genotypeAliasFR19_H 4.4358 -29.7473 33.8558 -1.8
## genotypeAliasH99_H 3.2519 -47.0806 24.3061 -7.8
## genotypeAliasHMV5301_H 137.5422 72.9638 173.1583 -3.4
## genotypeAliasHMV5325_H 6.5179 -13.0006 31.6095 4.1
## genotypeAliasHMV5343_H -44.8345 -72.8181 -21.8152 -0.5
## genotypeAliasHMV5347_H 13.0434 -9.0695 46.6929 0.6
## genotypeAliasHMV5405_H 66.7004 43.8041 96.0180 -2.2
## genotypeAliasHMV5409_H -58.1978 -85.3125 -37.1883 -2.9
## genotypeAliasHMV5422_H 32.6197 9.1898 58.0357 3.9
## genotypeAliasHMV5502_H -71.4391 -108.4259 -44.0211 -4.7
## genotypeAliasI198_H 21.3798 -19.2664 59.5909 -1.9
## genotypeAliasI205_H -120.5309 -156.3450 -87.8246 1.4
## genotypeAliasI211_H 25.6355 -4.0933 43.6974 2.2
## genotypeAliasI233_H -23.1715 -50.5330 1.5354 -0.8
## genotypeAliasI238_H -89.2717 -126.1546 -50.6716 -1.4
## genotypeAliasI242_H -35.4572 -73.0912 3.3589 0.0
## genotypeAliasI261_H -23.7325 -52.1576 -2.8900 3.4
## genotypeAliasI267_H 47.5455 22.7703 94.8904 0.3
## genotypeAliasIDT_H 3.8472 -40.0197 45.8599 -2.5
## genotypeAliasLAN496_H 0.6580 -49.7522 17.8352 0.0
## genotypeAliasLH123Ht_H 44.4655 18.5920 79.5700 0.5
## genotypeAliasLH145_H -70.7817 -126.0444 -24.1837 -1.3
## genotypeAliasLH38 -153.4844 -177.0984 -116.9854 2.5
## genotypeAliasLH38_H 40.8373 1.5949 69.2436 1.7
## genotypeAliasLH39_H 14.7957 -26.6671 53.0947 -2.5
## genotypeAliasLH59_H -17.9935 -59.2416 31.4904 -2.5
## genotypeAliasLH60_H -30.6832 -80.3048 1.2492 -6.5
## genotypeAliasLH65_H -34.3487 -69.3386 12.9737 -1.4
## genotypeAliasLH74_H -89.9754 -134.8108 -64.6637 -0.4
## genotypeAliasLH82_H -19.6203 -46.2276 12.7282 0.1
## genotypeAliasLH93_H 29.9592 -26.5677 54.6645 -5.6
## genotypeAliasLo1016_H 34.2114 -2.3664 93.5648 -1.3
## genotypeAliasLo1026_H 5.6793 -14.6983 44.3973 1.1
## genotypeAliasLo1035_H -28.3909 -49.7111 -8.5025 -1.4
## genotypeAliasLo1038_H -45.6067 -95.2249 -13.1987 1.0
## genotypeAliasLo1056_H 24.2675 -3.3063 42.9484 -1.6
## genotypeAliasLo1063_H -30.9128 -52.9641 -3.9237 4.6
## genotypeAliasLo1087_H -87.5644 -110.5211 -62.6698 2.9
## genotypeAliasLo1094_H 37.9743 10.6036 65.4601 -0.4
## genotypeAliasLo1095_H 21.1853 -0.3556 51.3739 4.0
## genotypeAliasLo1101_H -38.1653 -83.2291 -1.6888 -3.7
## genotypeAliasLo1106_H -51.8947 -74.3595 -23.4128 1.8
## genotypeAliasLo1123_H 21.7625 -16.4428 73.3661 -1.4
## genotypeAliasLo1124_H 6.4539 -13.9844 29.3269 10.7
## genotypeAliasLo1130_H -30.5914 -61.0278 2.4594 -3.4
## genotypeAliasLo1172_H 99.5022 76.0300 127.6428 0.8
## genotypeAliasLo1180_H 32.2811 11.6327 67.3551 -0.2
## genotypeAliasLo1187_H -37.0427 -76.3774 -16.7685 -1.9
## genotypeAliasLo1199_H -27.2459 -51.9899 -8.5974 1.3
## genotypeAliasLo1203_H -19.8831 -43.6734 18.7033 -1.7
## genotypeAliasLo1223_H 36.6878 20.3723 61.2374 1.1
## genotypeAliasLo1242_H -11.7999 -37.4610 17.4032 5.9
## genotypeAliasLo1251_H -4.1314 -43.1800 17.0810 -2.7
## genotypeAliasLo1253_H 2.5724 -32.1332 44.7378 -1.6
## genotypeAliasLo1261_H -43.3077 -73.7736 -11.0536 2.9
## genotypeAliasLo1266_H 59.7490 1.5616 89.9647 2.3
## genotypeAliasLo1270_H 56.2168 22.8268 80.3427 0.7
## genotypeAliasLo1273_H 48.8018 25.7510 110.9303 -0.5
## genotypeAliasLo1274_H 117.1753 81.0150 138.2785 -0.6
## genotypeAliasLo1280_H 54.8899 -10.2374 91.8791 -7.3
## genotypeAliasLo1282_H 47.6600 11.0182 68.8865 -2.1
## genotypeAliasLo1284_H 21.8834 -29.9086 48.1765 -1.9
## genotypeAliasLo1288_H -45.8825 -76.9416 -17.3575 0.1
## genotypeAliasLo1290_H 77.0138 52.6216 105.5618 2.2
## genotypeAliasLo1301_H -3.0395 -33.9849 40.4051 -0.5
## genotypeAliasLo904_H -48.7885 -64.3949 -21.4116 0.9
## genotypeAliasLp5_H -11.7427 -48.4239 30.8460 -0.5
## genotypeAliasML606_H 41.2114 -3.4566 75.8595 0.3
## genotypeAliasMo15W_H 29.1897 -2.4116 54.2159 -2.6
## genotypeAliasMo17 -311.3684 -348.9074 -268.7304 2.8
## genotypeAliasMo17_H 59.4307 17.8201 90.1305 1.3
## genotypeAliasMS153 -121.9660 -159.1261 -86.5114 3.2
## genotypeAliasMS153_H 48.2897 24.8366 86.1983 2.2
## genotypeAliasMS153_SILK 38.6684 7.3362 65.3203 -0.7
## genotypeAliasMS71_H -29.9832 -65.2146 4.9583 -2.5
## genotypeAliasN16_H 4.9523 -32.6605 45.1150 0.1
## genotypeAliasN192_H -32.9331 -56.1907 -9.8912 1.9
## genotypeAliasN22_H -67.5221 -105.8310 -46.4097 -9.5
## genotypeAliasN25_H -46.7615 -69.9110 -17.5418 0.0
## genotypeAliasN6_H -22.4501 -70.6346 6.9199 -0.2
## genotypeAliasNC290_H -41.8737 -82.0667 -26.1801 1.4
## genotypeAliasNC358_H -52.6064 -91.5945 -21.7539 3.7
## genotypeAliasNDB8_H 42.6665 5.3934 78.4731 -0.5
## genotypeAliasNK764_H -27.3230 -63.5093 -7.2848 -0.5
## genotypeAliasNK807_H -4.3148 -40.7752 34.7281 -1.8
## genotypeAliasNQ508_H -0.0257 -49.4275 30.8960 0.2
## genotypeAliasNS501_H 14.9110 -26.0980 40.1399 0.2
## genotypeAliasNS701_H 38.3863 -4.7684 82.5674 -2.3
## genotypeAliasOh02_H 8.4133 -17.7441 45.2999 5.4
## genotypeAliasOh33_H 9.0987 -24.5244 37.5531 -1.2
## genotypeAliasOh40B_H -32.4969 -56.8538 -17.5267 2.2
## genotypeAliasOh43 -207.9746 -236.1118 -151.4323 8.2
## genotypeAliasOh43_H 137.6866 97.1757 170.0401 6.0
## genotypeAliasOh43_SILK 62.9806 25.4393 83.5422 -4.1
## genotypeAliasOs426_H -58.0389 -76.4178 -25.7680 3.6
## genotypeAliasP465P_H -24.3036 -77.1944 16.6073 -1.2
## genotypeAliasPa35_H -55.1760 -97.7421 -30.5173 -0.9
## genotypeAliasPa36_H -15.7666 -41.7581 8.4925 1.1
## genotypeAliasPa374_H -13.4556 -48.2506 39.6466 -0.9
## genotypeAliasPa405_H -10.9355 -39.2599 21.7862 -1.0
## genotypeAliasPa91_H 79.0610 34.4282 124.5759 -4.7
## genotypeAliasPB116_H 31.9436 -10.5747 54.4968 -4.9
## genotypeAliasPB98TR_H -29.5586 -60.5039 -8.0206 -5.0
## genotypeAliasPH207_H -93.3195 -127.1792 -71.9081 -1.6
## genotypeAliasPHB09_H -46.7440 -79.0350 -20.1476 -6.8
## genotypeAliasPHG35_H 60.6127 6.1538 82.8289 -3.5
## genotypeAliasPHG39_H 28.2011 -7.1656 59.7728 -1.7
## genotypeAliasPHG47_H 82.1234 10.0487 116.0044 -4.3
## genotypeAliasPHG50_H 59.7727 31.6362 100.0496 0.2
## genotypeAliasPHG71_H 21.9966 -25.1502 52.7295 -2.2
## genotypeAliasPHG80_H -53.9288 -95.1778 -20.7154 0.4
## genotypeAliasPHG83_H -73.0015 -122.4170 -51.9958 0.2
## genotypeAliasPHG84_H 90.0107 68.0902 118.5824 -1.4
## genotypeAliasPHG86_H -20.3597 -73.7470 7.3174 -2.8
## genotypeAliasPHH93_H 43.9510 -18.2044 69.9202 -5.9
## genotypeAliasPHJ40_H 94.4187 70.6233 115.8367 2.6
## genotypeAliasPHK29_H 40.6430 12.1787 86.6364 -0.8
## genotypeAliasPHK76_H 5.6263 -18.3114 39.2293 -0.8
## genotypeAliasPHR36_H 96.0153 37.4911 138.8841 -4.5
## genotypeAliasPHT77_H 59.3523 42.3303 86.8147 1.0
## genotypeAliasPHV63_H 56.4534 28.0692 81.7999 -2.8
## genotypeAliasPHW65_H -0.7724 -44.9244 24.3476 -5.9
## genotypeAliasPHZ51_H 117.2205 87.8109 149.9307 -1.1
## genotypeAliasPP147_H 21.3939 -32.8841 54.2785 1.8
## genotypeAliasSC-Malawi_H -10.1327 -38.6521 17.7138 0.6
## genotypeAliasUH_2500_H -6.3234 -34.6501 37.2207 -0.5
## genotypeAliasUH250_H 70.8540 40.8624 96.9486 -0.7
## genotypeAliasUH304_H -46.4057 -105.7144 -16.6218 -2.6
## genotypeAliasUH_6102_H 24.2133 2.7987 56.3390 -0.3
## genotypeAliasUH_6179_H -51.0781 -87.7474 -24.2470 -1.6
## genotypeAliasUH_P024_H -132.9016 -157.4648 -93.0016 4.2
## genotypeAliasUH_P060_H 37.8416 -0.1161 77.6244 -4.1
## genotypeAliasUH_P064_H 12.8759 -23.6575 34.2524 2.4
## genotypeAliasUH_P074_H -65.5361 -98.2608 -43.0155 2.8
## genotypeAliasUH_P087_H -36.2252 -56.0842 -6.4146 1.0
## genotypeAliasUH_P089_H 17.0583 -3.6671 46.8271 -1.9
## genotypeAliasUH_P104_H 103.0462 65.4691 132.6336 -4.5
## genotypeAliasUH_P115_H 85.5897 61.1030 111.4937 0.4
## genotypeAliasUH_P128_H 78.8203 50.5082 113.9136 -1.9
## genotypeAliasUH_P148_H -30.2066 -64.6634 -3.2706 -2.2
## genotypeAliasUH_S018_H -8.3702 -28.9768 12.2129 -1.5
## genotypeAliasUH_S020_H -45.7264 -97.2854 -13.8611 -2.7
## genotypeAliasUH_S025_H 82.7534 49.9840 108.8224 -2.6
## genotypeAliasVa26_H 43.8857 19.3223 75.1117 2.9
## genotypeAliasW117 -144.4556 -194.3770 -86.2115 3.7
## genotypeAliasW117_H 128.1677 86.0450 173.1891 2.1
## genotypeAliasW153R_H -45.7736 -86.4146 -20.0249 -5.3
## genotypeAliasW182E_H -14.7338 -31.4863 6.9002 0.5
## genotypeAliasW23_H 17.5847 -23.1603 49.1260 -2.7
## genotypeAliasW602S_H 26.3704 -3.3642 49.9855 -1.6
## genotypeAliasW604S_H -51.7390 -88.5164 -24.1534 0.5
## genotypeAliasW64A_H -60.4985 -86.3283 -36.6257 -0.6
## genotypeAliasW9 -474.3791 -533.2429 -433.1502 0.0
## genotypeAliasW95115_H 31.1635 -15.3837 49.6413 -1.1
## genotypeAliasW9_H -39.7016 -103.1672 4.9525 1.8
## genotypeAliasWf9_H 139.4442 124.3776 170.3892 0.8
## genotypeAliasX061_H 60.9865 36.8142 88.0306 -1.4
## genotypeAliasX697_H 33.9312 5.0678 61.6646 -1.9
## tau2.S 4625.3717 2713.7318 5144.0853 -5.6
## tau2.T 11305.8163 2148.5654 45679.4499 20.5
## nu2 6303.3624 6165.8336 9365.4338 2.5
## rho.S 0.4136 0.2237 0.4799 -7.8
## rho.T 0.9099 0.6090 0.9999 -18.1
## DIC p.d WAIC p.w LMPL
## 496887.000 2147.925 504149.917 6642.962 -249609.718
## loglikelihood
## -246295.575outtmp<-outlierCARBayesST(modelin=model[[1]],datain=model[[2]],threshold=4,trait="plantHeight")
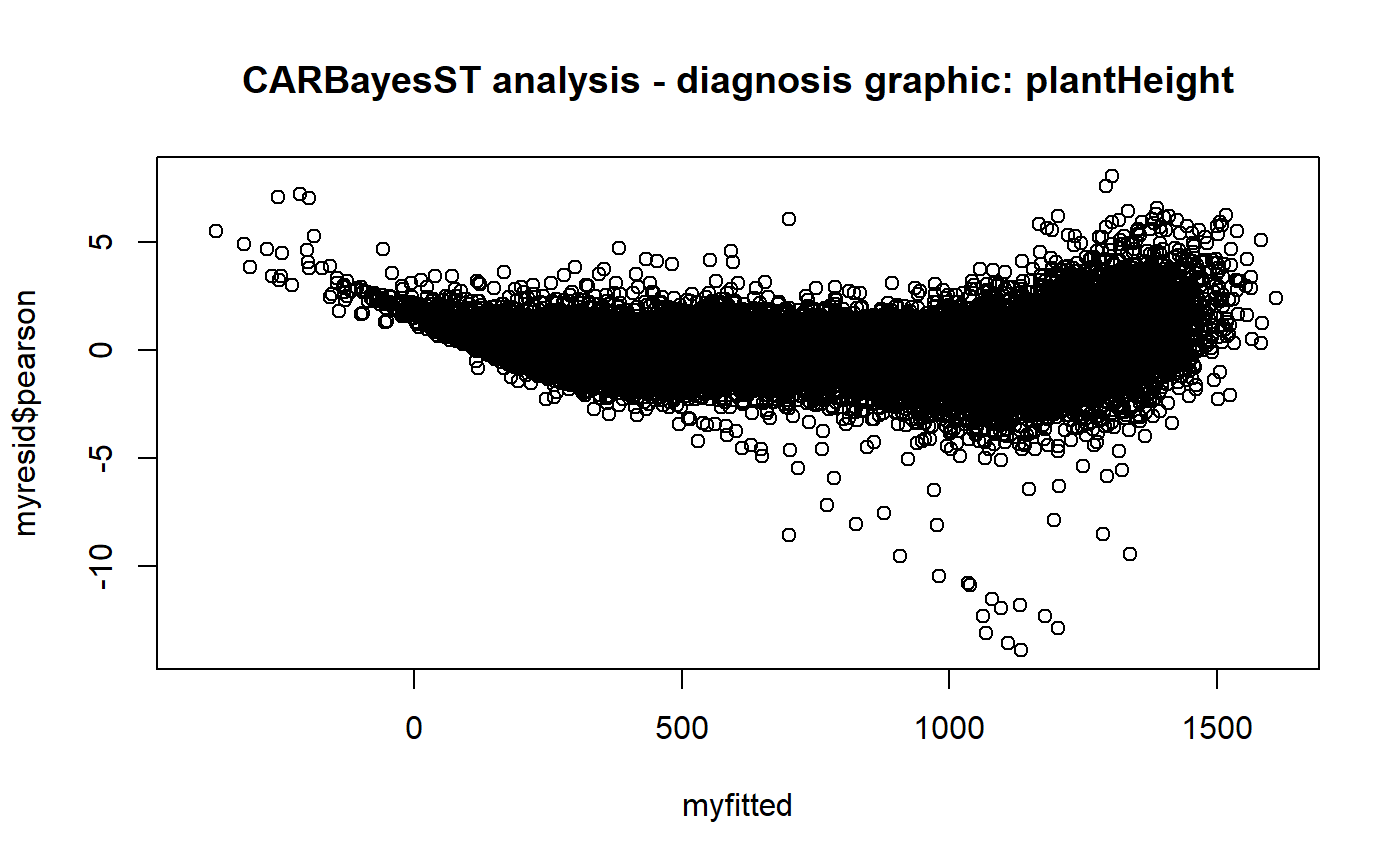
Outlier detection
The output report can be over-sized (more than 1Mb), for size of sub-directories in packages purposes, I choose to represent only the first genotypes…
mygeno<-as.character(unique(model[[2]][,"genotypeAlias"])) mygeno<-mygeno[1:6] for (i in seq(1,length(mygeno),by=15)){ myvec<-seq(i,i+14,1) plotCARBayesST(datain=model[[2]],outlierin=outtmp,myselect=myvec,trait="plantHeight",xvar="thermalTime") }
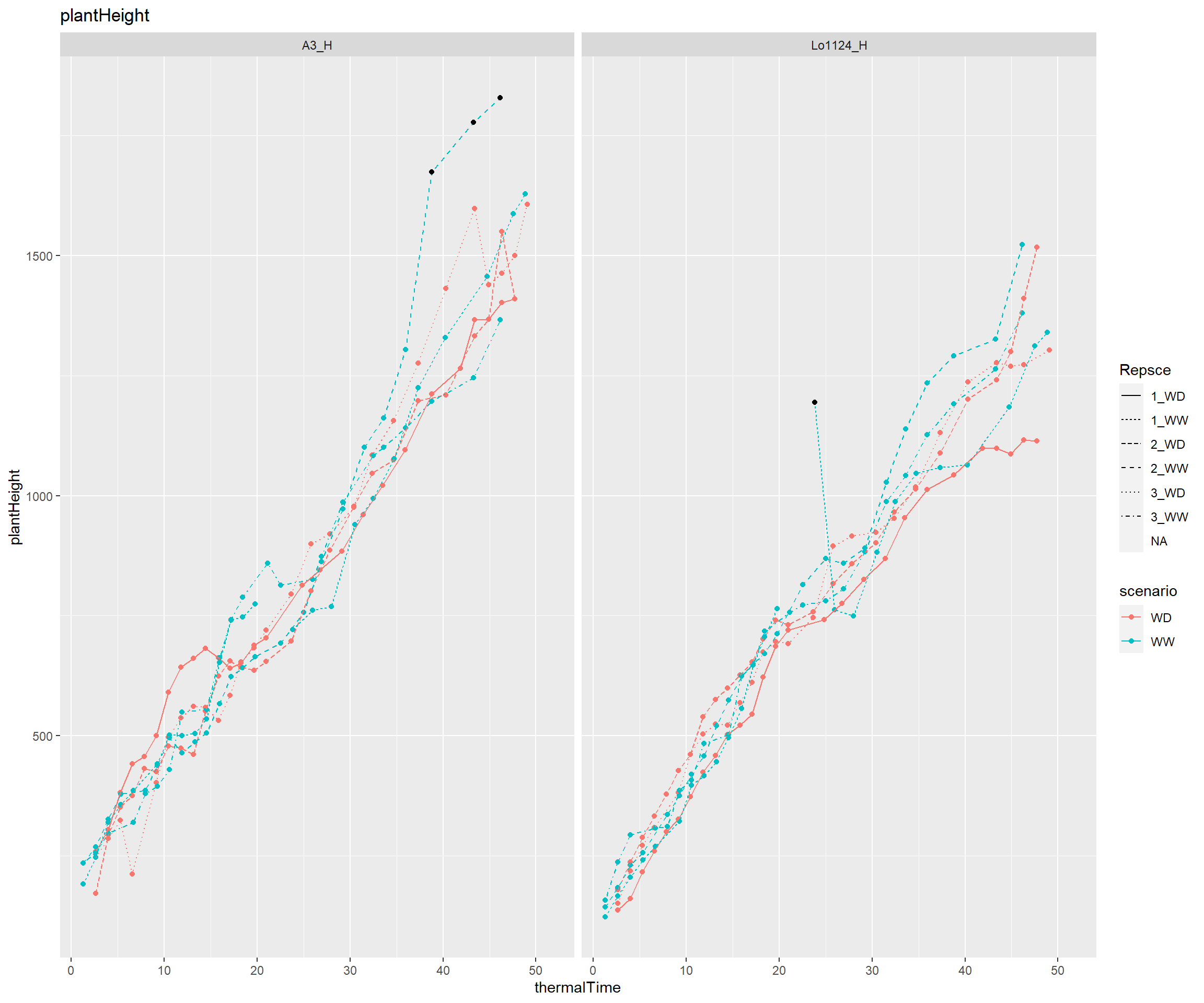
Session info
## R version 4.0.2 (2020-06-22)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 18363)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=French_France.1252 LC_CTYPE=French_France.1252
## [3] LC_MONETARY=French_France.1252 LC_NUMERIC=C
## [5] LC_TIME=French_France.1252
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] openSilexStatR_1.1.0 dplyr_1.0.2 lubridate_1.7.9
##
## loaded via a namespace (and not attached):
## [1] tidyr_1.1.2 splines_4.0.2 dotCall64_1.0-0 gtools_3.8.2
## [5] assertthat_0.2.1 expm_0.999-5 CARBayesdata_2.2 sp_1.4-2
## [9] stats4_4.0.2 yaml_2.2.1 LearnBayes_2.15.1 truncdist_1.0-2
## [13] pillar_1.4.6 backports_1.1.9 lattice_0.20-41 glue_1.4.2
## [17] digest_0.6.25 RColorBrewer_1.1-2 colorspace_1.4-1 plyr_1.8.6
## [21] htmltools_0.5.0 Matrix_1.2-18 pkgconfig_2.0.3 raster_3.3-13
## [25] CARBayesST_3.1 gmodels_2.18.1 purrr_0.3.4 scales_1.1.1
## [29] gdata_2.18.0 tibble_3.0.3 farver_2.0.3 generics_0.0.2
## [33] ggplot2_3.3.2 ellipsis_0.3.1 magrittr_1.5 crayon_1.3.4
## [37] deldir_0.1-28 memoise_1.1.0 evaluate_0.14 GGally_2.0.0
## [41] fs_1.4.2 nlme_3.1-148 MASS_7.3-51.6 foreign_0.8-80
## [45] truncnorm_1.0-8 class_7.3-17 data.table_1.13.0 tools_4.0.2
## [49] shapefiles_0.7 lifecycle_0.2.0 matrixStats_0.56.0 stringr_1.4.0
## [53] munsell_0.5.0 compiler_4.0.2 pkgdown_1.5.1 e1071_1.7-3
## [57] evd_2.3-3 rlang_0.4.7 classInt_0.4-3 units_0.6-7
## [61] grid_4.0.2 rstudioapi_0.11 htmlwidgets_1.5.1 spam_2.5-1
## [65] crosstalk_1.1.0.1 labeling_0.3 rmarkdown_2.3 SpATS_1.0-11
## [69] boot_1.3-25 testthat_2.3.2 gtable_0.3.0 codetools_0.2-16
## [73] reshape_0.8.8 DBI_1.1.0 R6_2.4.1 gridExtra_2.3
## [77] knitr_1.29 rgdal_1.5-16 rprojroot_1.3-2 spdep_1.1-5
## [81] KernSmooth_2.23-17 desc_1.2.0 matrixcalc_1.0-3 stringi_1.4.6
## [85] Rcpp_1.0.5 vctrs_0.3.4 sf_0.9-5 leaflet_2.0.3
## [89] spData_0.3.8 tidyselect_1.1.0 xfun_0.16 coda_0.19-3References
- R Development Core Team (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org.
- Roger S. Bivand, Edzer Pebesma, Virgilio Gomez-Rubio, 2013. Applied spatial data analysis with R, Second edition. Springer, NY. http://www.asdar-book.org/
- Duncan Lee, Alastair Rushworth and Gary Napier (2017). CARBayesST: Spatio-Temporal Generalised Linear Mixed Models for Areal Unit Data. R package version 2.5. https://CRAN.R-project.org/package=CARBayesST